Abstract
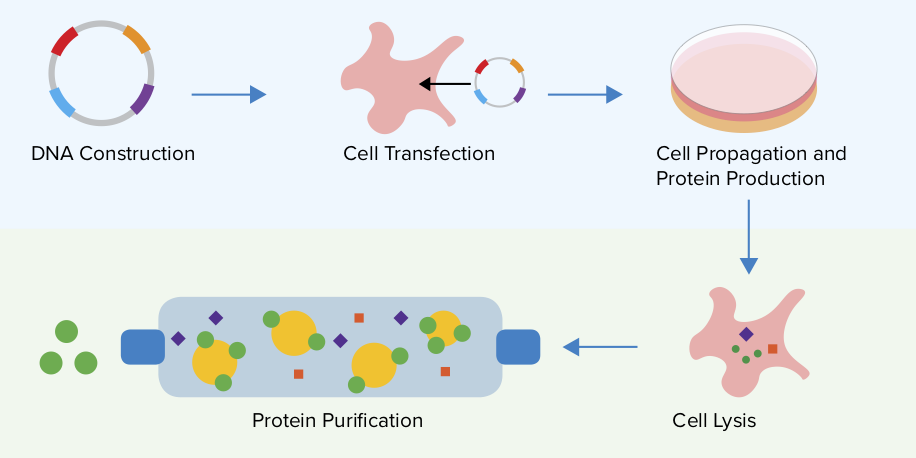
Pure, soluble and functional proteins are in great demand in modern biotechnology. Natural protein sources rarely meet the requirements for quantity, ease of isolation, or price, and therefore recombinant technology is often the method of choice. Recombinant cell factories are constantly in use for the production of protein preparations for further purification and processing.
Escherichia coli is a frequently used host as it facilitates protein expression due to its relative simplicity, low cost and rapid high-density culture, well-known genetics, and a large number of compatible molecular tools available. Despite all of these qualities, the expression system of recombinant proteins with E. coli as the host often results in insoluble and/or non-functional proteins. Here we review new approaches to overcome these obstacles by strategies that focus on the controlled expression of the target protein in an unmodified form or by applying modifications via expressivity and solubility tags.
Interaction partners and protein folding.
The insolubility of the protein in the E. coli cytoplasm is partially related to the distribution of hydrophobic residues on the protein surface. Therefore, soluble expression of heteromultimeric protein subunits sometimes suffers from inclusion body formation in the absence of a suitable binding partner. Soluble expression in E. coli of the bacteriophage T4 gene 23 product (major capsid protein) required co-expression of the gene 31 product (phage co-chaperonin gp31). Expression of the correct interaction partner allowed gp23 to fold correctly and form long regular structures in the E. coli cytoplasm.
Another study reports the purification of a heterodimeric complex by expression of each subunit (pheromazein A and C) as a fusion to thioredoxin. Each subunit remained soluble in solution, when thioredoxin was proteolytically removed, only in the presence of the other. In conclusion, interaction partners potentially favour the in vivo solubility of target proteins. The new systems for the coexpression of multiple proteins involved in complex structures allow this type of strategy.
E. coli expression system
Escherichia coli (E. coli) is one of the most widely used hosts for the production of heterologous proteins and its genetics is much better characterized than that of any other microorganism. Recent advances in the fundamental understanding of protein transcription, translation, and folding in E. coli, coupled with serendipitous discoveries and the availability of improved genetic tools, make this bacterium more valuable than ever for the expression of complex eukaryotic proteins.
The following factors or technical approaches are generally considered for successful recombinant protein expression in E. coli:
1. Initial Expression Screening
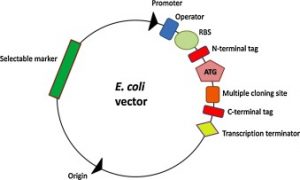
To clone the gene of interest into a variety of E. coli expression vectors with different expression tags or fusion proteins and express them in a basic strain of E. coli. To clone the gene of interest into an E. coli regular expression vector and express it in a variety of E. coli host strains.
2. Optimization of expression levels
Examine the codon usage of the heterologous protein. The following problems are often encountered:
- Examining the second codon.
- Minimization of GC content at the 5′ end. Addition of a transcription terminator (or an additional one if one is already present).
- Adding a merging partner.
- Use of protease-deficient host strains.
3. Improved protein solubility
- Reduce the rate of protein synthesis.
- Change the culture medium
- Co-expression of chaperones and/or foldases
- Periplasmic expression
- Use of specific host strains
- Adding a merging partner
- Expression of a protein fragment.
- In vitro denaturation and protein refolding
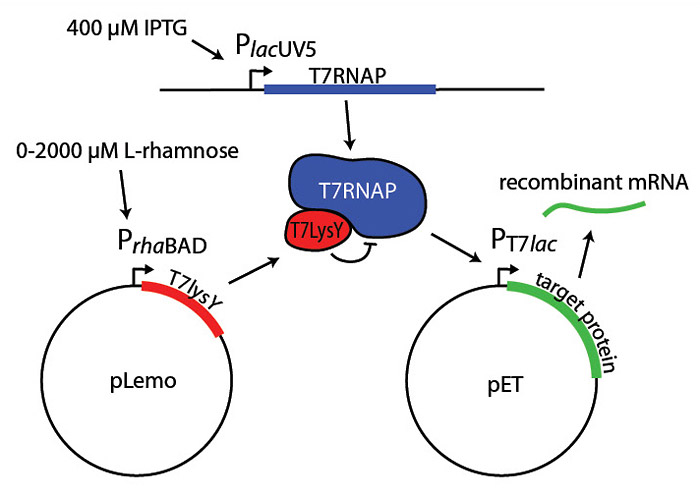
4. Improved protein stability
- N-terminal rule: In E. coli, the N-terminal residues Arg, Lys, Leu, Phe, Tyr, and Trp greatly reduce the half-life of the protein.
- PEST hypothesis – In eukaryotes, proteins are destabilized by regions enriched in Pro, Glu, Ser and Thr.
- Use of protease-deficient host strains: The use of host strains that carry mutations that eliminate protease production can sometimes improve accumulation by reducing proteolytic degradation. BL21, the workhorse of E. coli expression, is deficient in two proteases encoded by the loan (cytoplasmic) and ompT (periplasmic) genes.
- Periplasmic expression: in the periplasm, the proteolytic degradation of proteins is reduced. This is mainly because the total number of proteins in the periplasm is less.
- Lowering the growth temperature: Reducing the growth temperature will result in slower protein production, but also slower proteolytic degradation.
5. In vitro expression using E. coli extracts
E. coli-based in vitro transcription/translation expression systems have been used for specific applications:
- Expression of toxic recombinant proteins.
- Labelling of recombinant proteins.
- Incorporation of unnatural amino acids.
- Production of small amounts of protein quickly and cheaply.